Part:BBa_K1062000:Design
Guide RNA (gRNA) target for RFP
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Design Notes
Part:BBa_K1062000 is a gRNA containing a CSY4 cut site and specifically targets the RFP gene.
gRNA
A gRNA is a 102 nt sequence in which contains: a 20 nt sequence which will contain the sequence of the intended target, a 42 nt Cas9-binding hairping, and a 40 nt transcription terminator.
Figure 1: An image show a general picture of a gRNA.
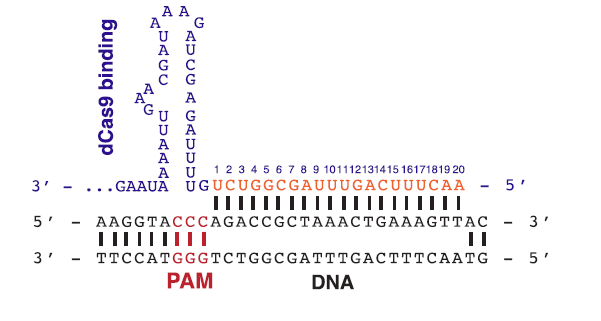
An important thing to note when using or creating a gRNA is that before the 20nt sequence, there must be a 3nt sequence called a PAM sequence which contain this sequence 5'-NGG-3'. This 3nt sequence is what the Cas9 use as a binding signal. It is also important to point out that the PAM sequence is never in the gRNA itself.
Figure 2: An example of a PAM sequence before the 20nt target sequence.
Csy4 Cutsite
The Csy4 cut site is a sequence that is essential to the creation of the gRNA.
The reason the Csy4 cut site to have the gRNA is because originally the gRNA would be in the plasmid before it can be used as a targeting system. However, in order to use the gRNA as a targeting system in CRISPRI, it needs to be cut away from the plasmid. To do this, an enzyme, when present, called Csy4 would cut the Csy4 cut site and free the gRNA from the plasmid.
Sources
The RFP gRNA was retrieved from Stanley Qi's Lab at UCSF.
References
Currently Stanley Qi's lab is doing research in which hopes to use the CRISPR system in new applications.
[http://www.ncbi.nlm.nih.gov/pubmed/23452860 Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression ]
In the paper in the link provided, Stanley had shown to control gene expression using the CRISPR system using GFP and RFP as proof of concept. The RFP gRNA being used in our project is the same gRNA Stanley used in this paper.